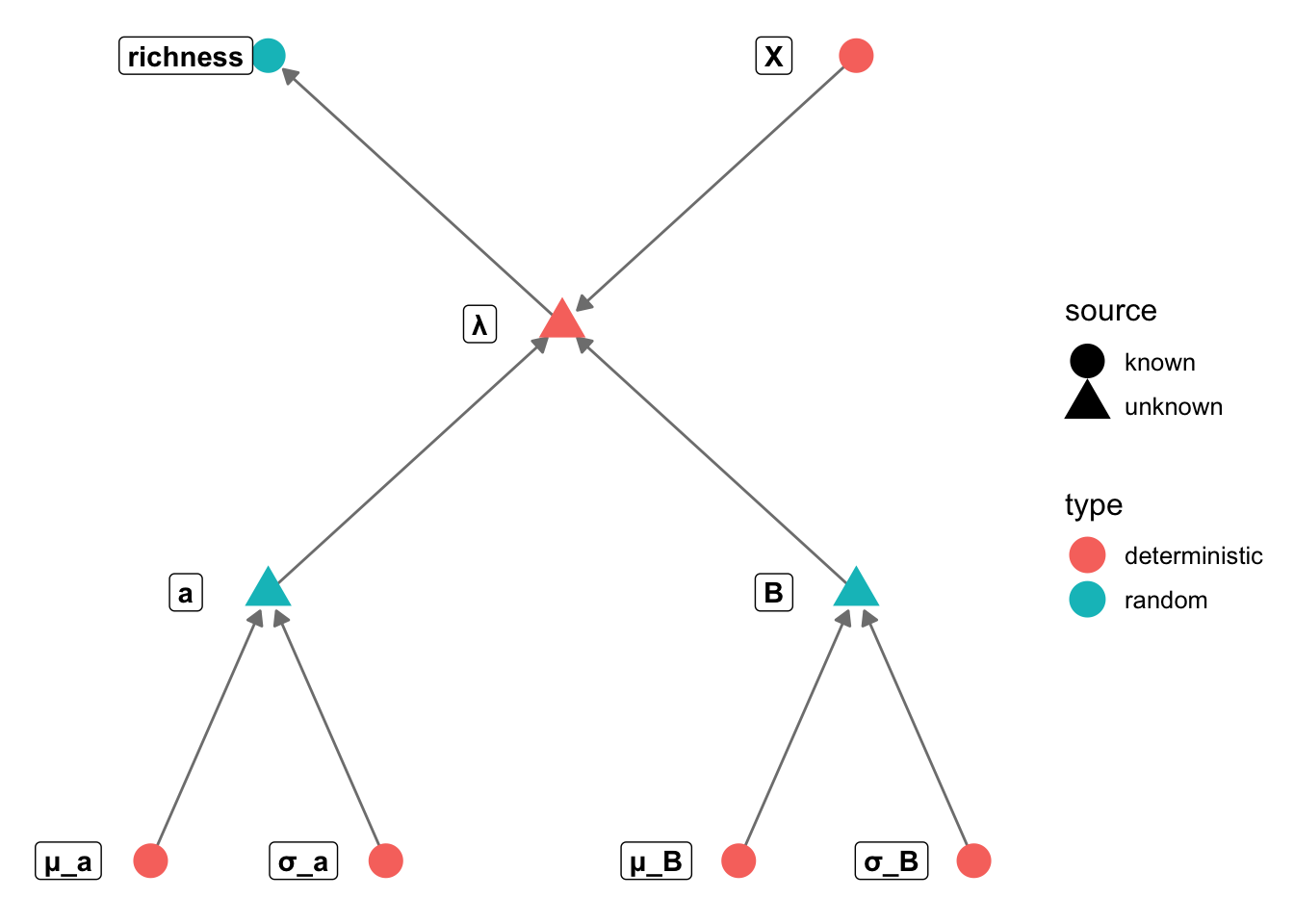
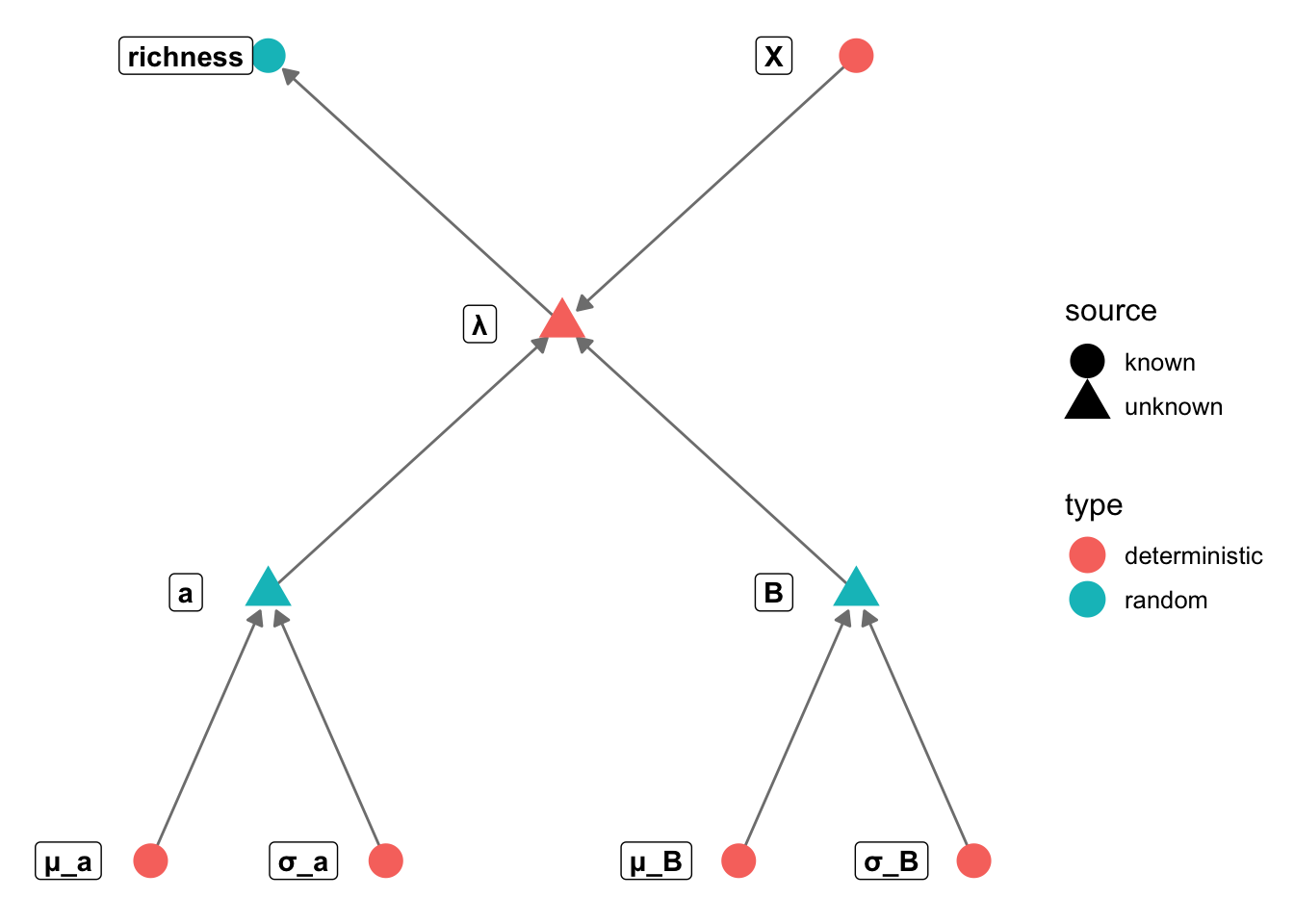
GLM in Stan
For this exercise, you will use the birddiv
(in vu_advstats_students/data/birddiv.csv) dataset; you can
load it directly from github using data.table::fread().
Bird diversity was measured in 1-km^2 plots in multiple countries of
Europe, investigating the effects of habitat fragmentation and
productivity on diversity. We will consider a subset of the data.
Specificially, we will ask how various covariates are associated with
the diversity of birds specializing on different habitat types. The data
have the following potential predictors:
- Grow.degd: growing degree days, a proxy for how
warm the climate is.
- For.cover: Forest cover near the sampling
location
- NDVI: normalized difference vegetation index, a
proxy for productivity
- For.diver: Forest diversity in the forested area
nearby
- Agr.diver: Diversity of agricultural
landscapes
- For.fragm: Index of forest fragmentation
All of the above variables are standardized to a 0-100 scale.
Consider this when choosing priors.
Your response variable will be richness, the bird
species richness in the plot. Additionally, you have an indicator
variable hab_type. This is not telling you what habitat
type was sampled (plots included multiple habitats). Rather, this is
telling you what type of bird species were counted for the richness
measurement: so hab_type == "forest" & richness == 7
indicates that 7 forest specialists were observed in that plot.
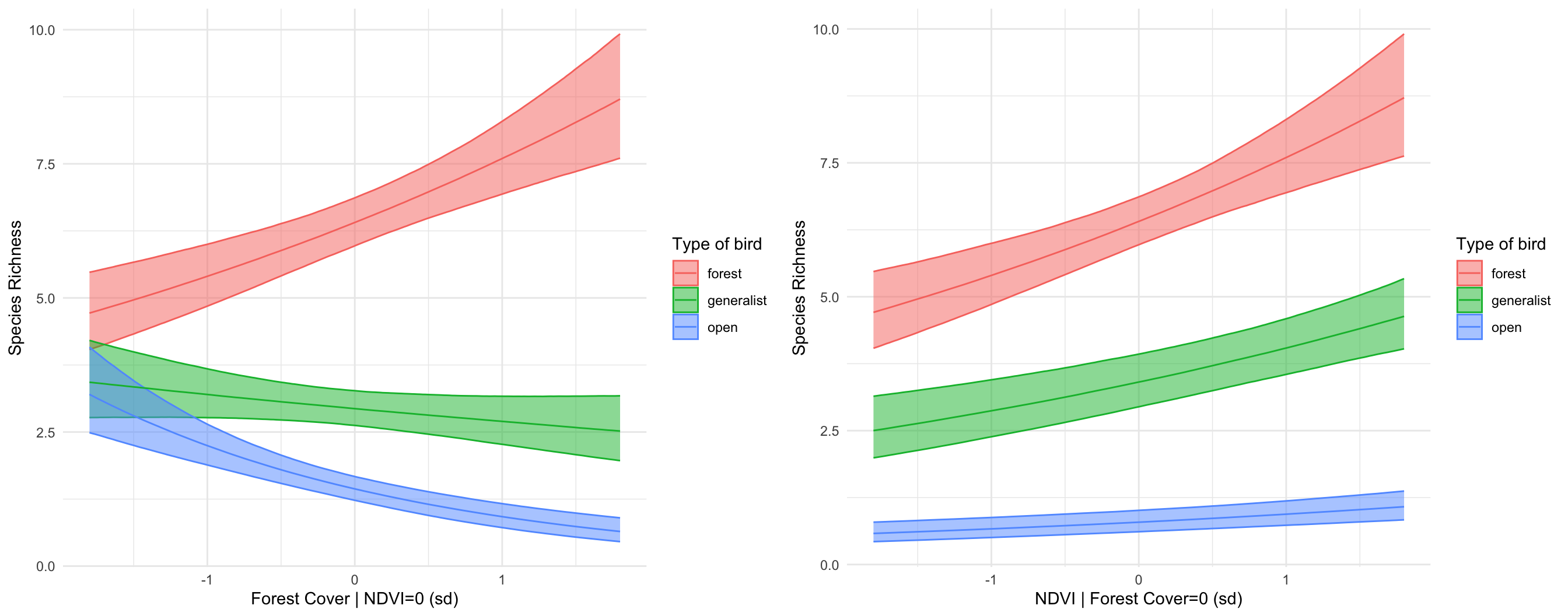
Build one or more generalised linear models for bird richness. Your
task should be to describe two things: (1) how does richness vary with
climate, productivity, fragmentation, or habitat diversity, and (2) do
these relationships vary depending on what habitat bird species
specialize on.
2. Sample from the joint posterior
First we compile the model and prepare the data.
library(rstan)
bird_div_glm = stan_model("stan/bird_div_glm.stan")
library(data.table)
birds = fread("../vu_advstats_students/data/birddiv.csv")
# stan can't handle NAs
birds = birds[complete.cases(birds)]
# turn predictors into a matrix
X = as.matrix(birds[, c("Grow.degd", "For.cover", "NDVI",
"For.diver", "Agr.diver", "For.fragm")])
# Remove the scale from X, make the model scale independent
X_scaled = scale(X)
2. Sample from the joint posterior
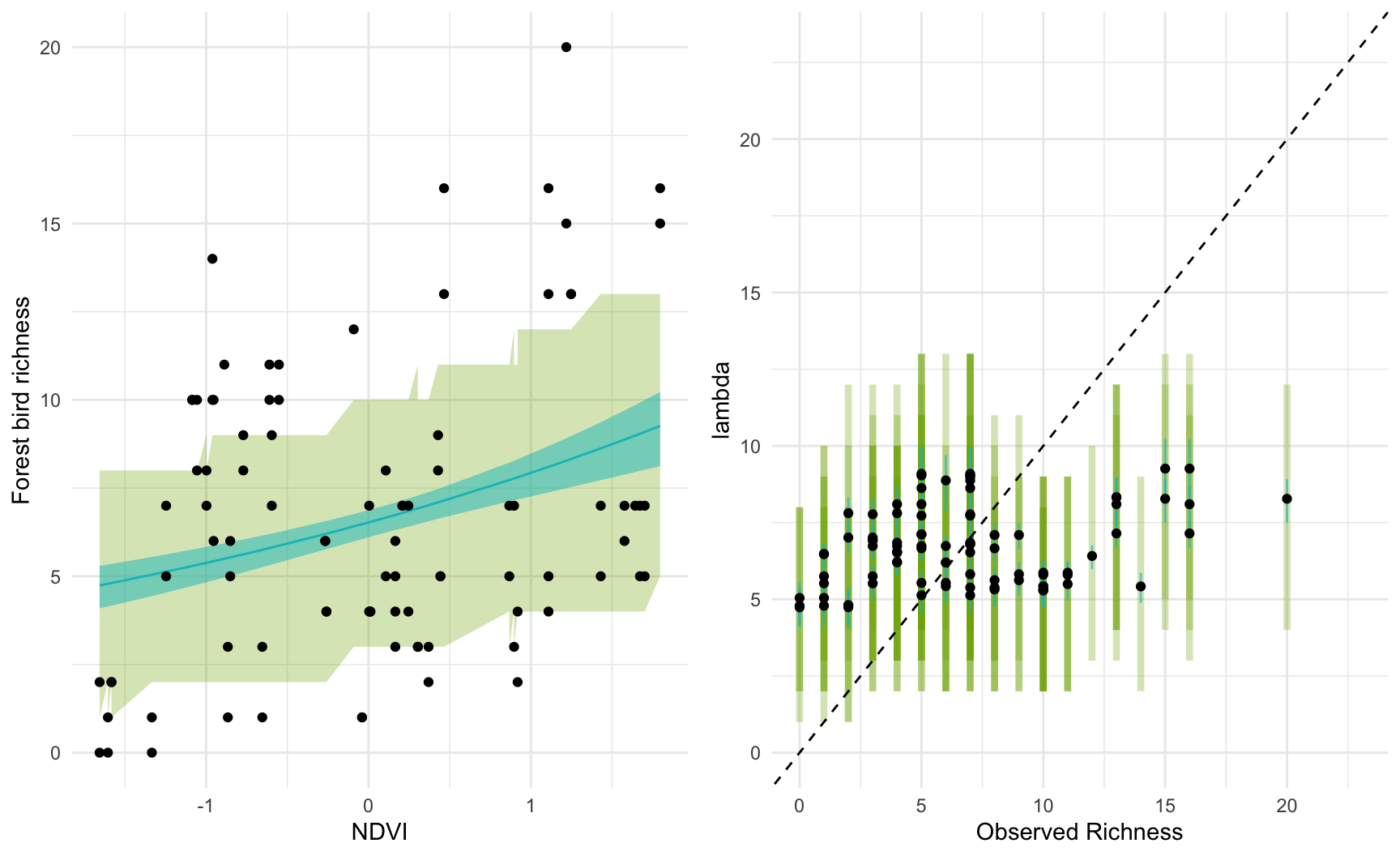
Next we fit an initial model using only NVDI as a predictor.
which_forest = which(birds$hab_type == "forest")
standat1 = list(
n = length(which_forest),
k = 1,
richness = birds$richness[which_forest],
X = X_scaled[which_forest, "NDVI", drop=FALSE],
mu_a = 0,
mu_b = 0,
# these prior scales are still SUPER vague
# exp(20) is a possible intercept under this prior!
sigma_a = 10,
sigma_b = 5
)
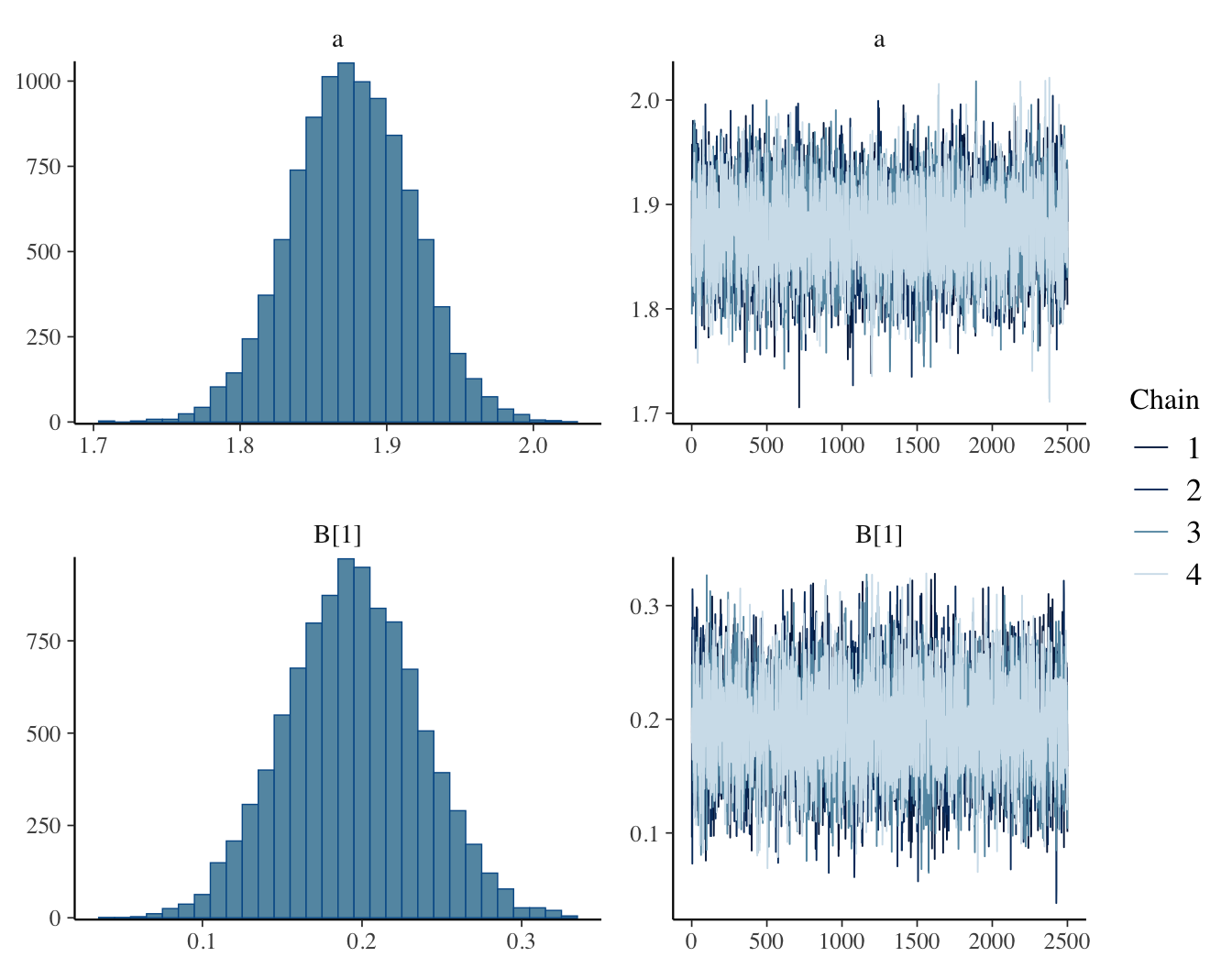
fit1 = sampling(bird_glm, data = standat1, iter=5000,
chains = 4, refresh = 0)
2. Sample from the joint posterior
## Inference for Stan model: anon_model.
## 4 chains, each with iter=5000; warmup=2500; thin=1;
## post-warmup draws per chain=2500, total post-warmup draws=10000.
##
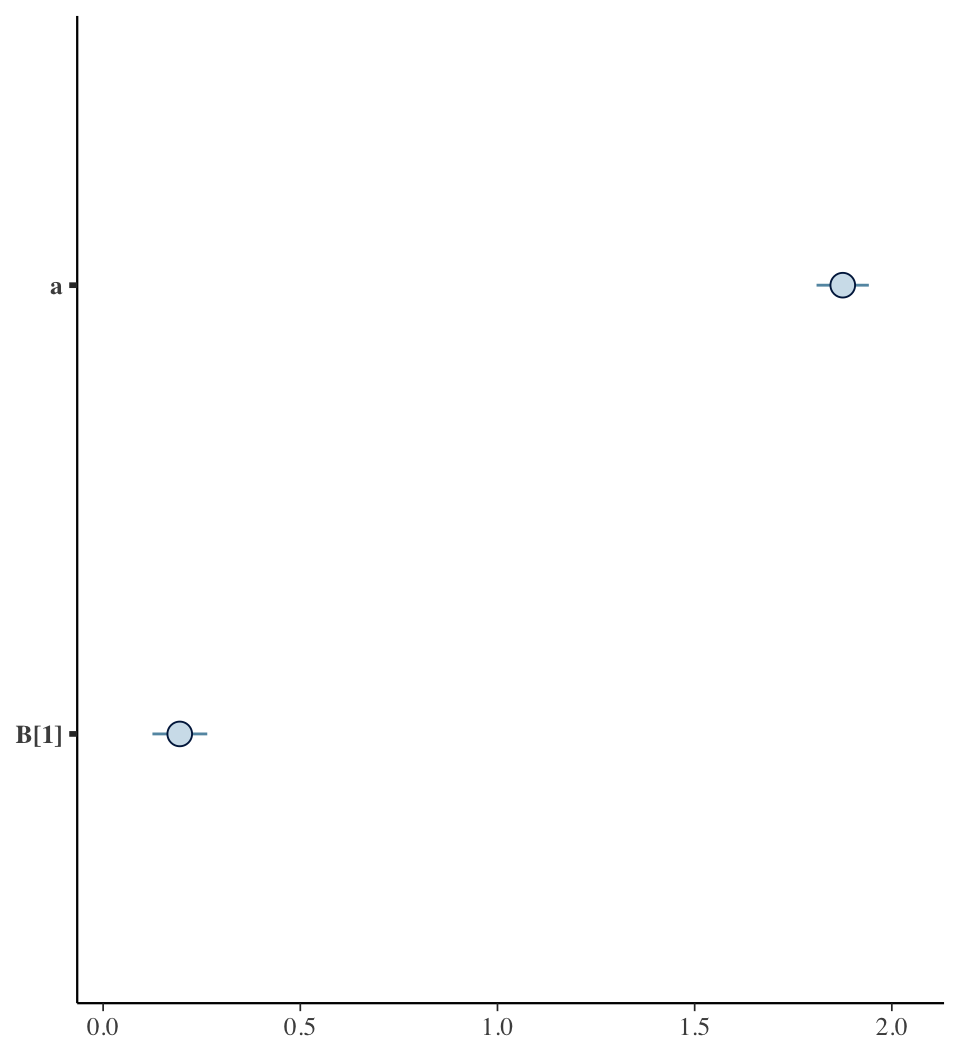
## mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
## a 1.88 0 0.04 1.80 1.85 1.88 1.90 1.96 6837 1
## B[1] 0.19 0 0.04 0.11 0.16 0.19 0.22 0.27 7036 1
##
## Samples were drawn using NUTS(diag_e) at Sun Jan 25 18:41:09 2026.
## For each parameter, n_eff is a crude measure of effective sample size,
## and Rhat is the potential scale reduction factor on split chains (at
## convergence, Rhat=1).
2. Sample from the joint posterior
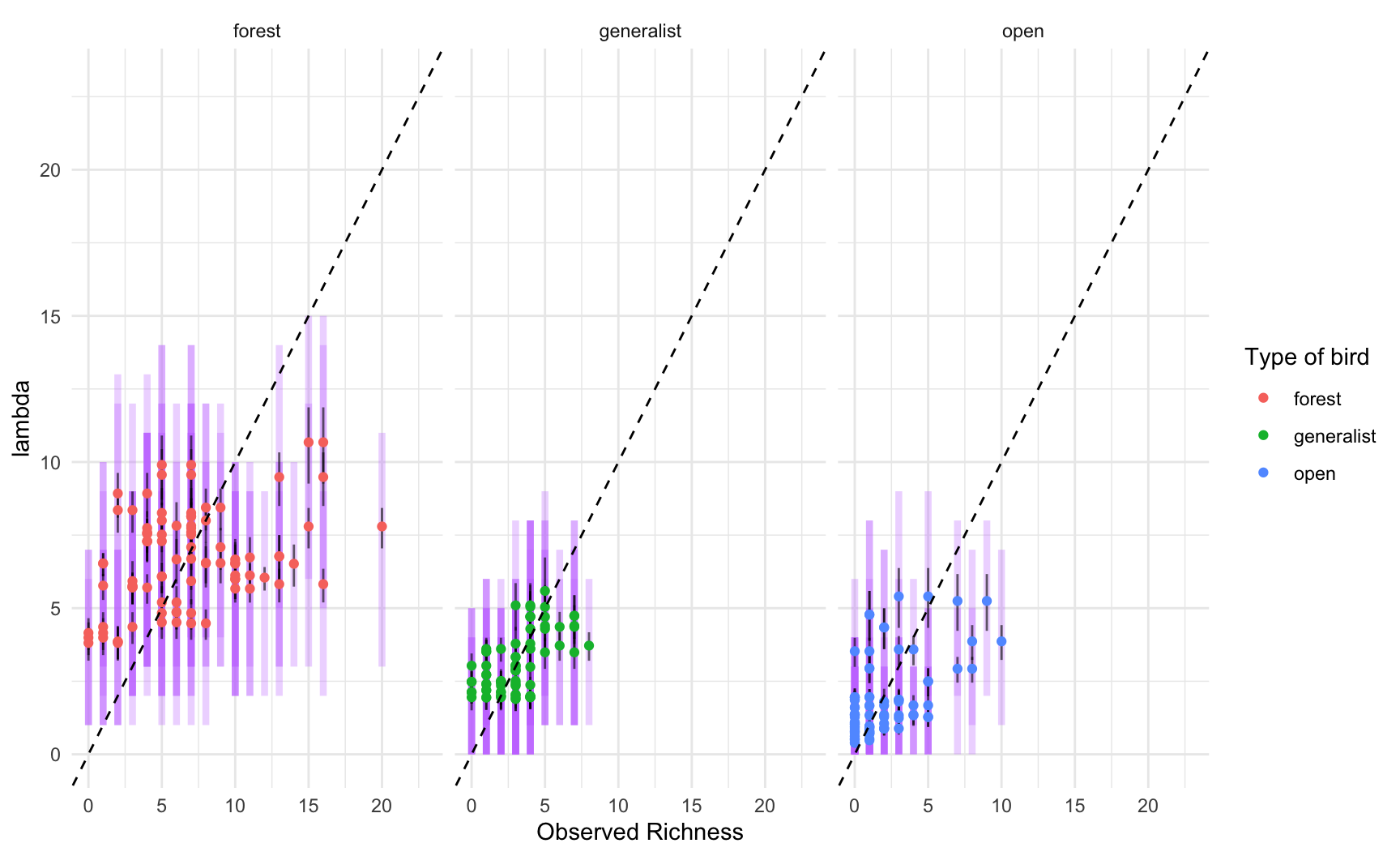
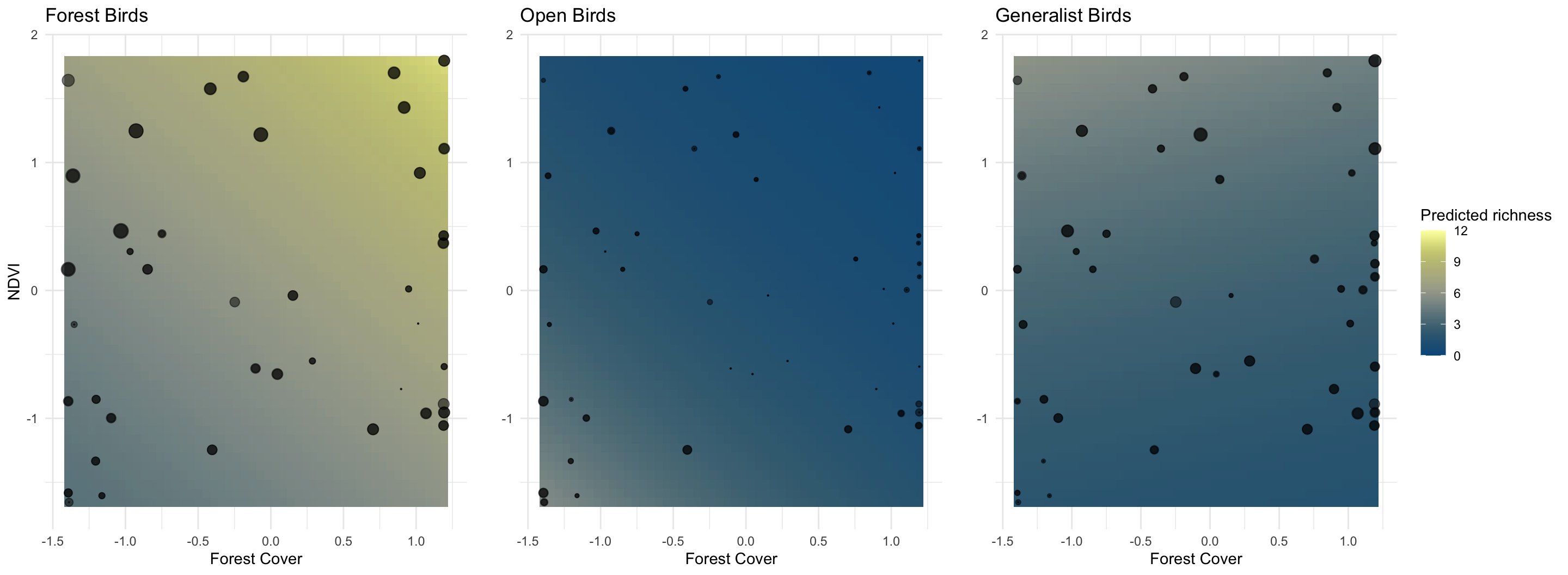
Next, we choose two variables, and try them using birds of different
habitat types.
# Second, looking at how two variables influence birds of different types
# grab two variables
X_2 = X_scaled[, c("For.cover", "NDVI")]
# add a categorical variable for bird type
X_2 = cbind(X_2, open=ifelse(birds$hab_type == "open",1, 0))
X_2 = cbind(X_2, generalist=ifelse(birds$hab_type == "generalist",1, 0))
# add interaction terms with the categories
X_2 = cbind(X_2,
op_forCov = X_2[,"For.cover"] * X_2[,"open"],
op_NDVI = X_2[, "NDVI"] * X_2[,"open"],
ge_forCov = X_2[,"For.cover"] * X_2[,"generalist"],
ge_NDVI = X_2[,"NDVI"] * X_2[,"generalist"])
head(X_2)
## For.cover NDVI open generalist op_forCov op_NDVI ge_forCov ge_NDVI
## [1,] 1.1895779 0.4284131 0 0 0 0 0 0
## [2,] -1.3937570 -0.8660731 0 0 0 0 0 0
## [3,] -0.4034797 -1.2463741 0 0 0 0 0 0
## [4,] -1.0985427 -0.9977157 0 0 0 0 0 0
## [5,] -1.3937570 -1.5827942 0 0 0 0 0 0
## [6,] 1.1930054 0.2090086 0 0 0 0 0 0
2. Sample from the joint posterior
Next, we choose two variables, and try them using birds of different
habitat types.
standat2 = list(
n = length(birds$richness),
k = ncol(X_2),
richness = birds$richness,
X = X_2,
mu_a = 0,
mu_b = 0,
# these prior scales are still SUPER vague (exp(20) is a possible intercept under this prior!)
sigma_a = 10,
sigma_b = 5
)
fit2 = sampling(bird_glm, data = standat2, iter=5000,
chains = 4, refresh = 0)
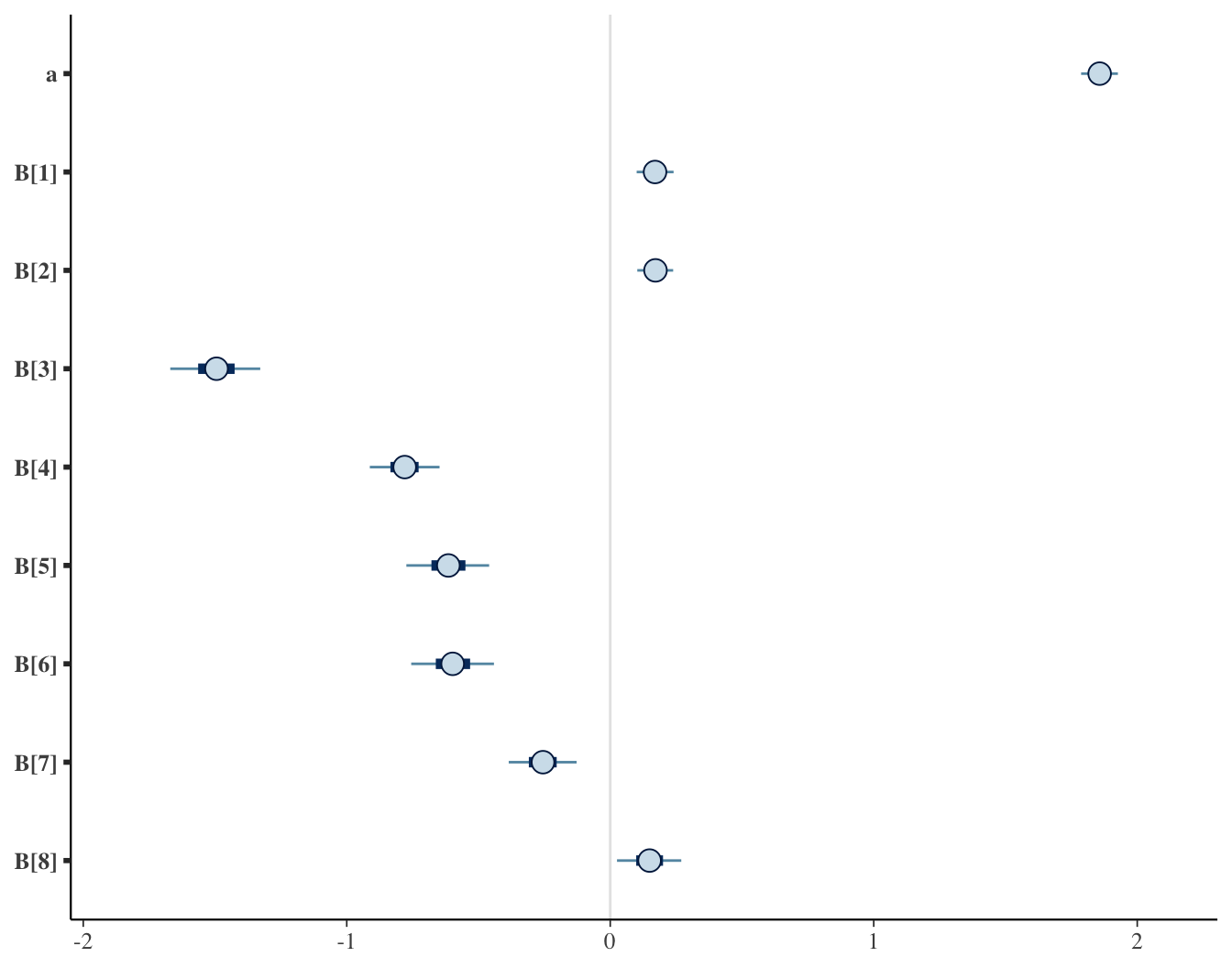
2. Sample from the joint posterior
## Inference for Stan model: anon_model.
## 4 chains, each with iter=5000; warmup=2500; thin=1;
## post-warmup draws per chain=2500, total post-warmup draws=10000.
##
## mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
## a 1.86 0 0.04 1.77 1.83 1.86 1.88 1.94 7822 1
## B[1] 0.17 0 0.04 0.09 0.14 0.17 0.20 0.25 7436 1
## B[2] 0.17 0 0.04 0.09 0.14 0.17 0.20 0.25 7963 1
## B[3] -1.49 0 0.10 -1.70 -1.56 -1.49 -1.42 -1.29 8848 1
## B[4] -0.78 0 0.08 -0.94 -0.83 -0.78 -0.72 -0.63 8091 1
## B[5] -0.61 0 0.10 -0.80 -0.68 -0.61 -0.55 -0.43 8522 1
## B[6] -0.60 0 0.10 -0.79 -0.66 -0.60 -0.53 -0.41 8654 1
## B[7] -0.26 0 0.08 -0.41 -0.31 -0.26 -0.20 -0.10 8876 1
## B[8] 0.15 0 0.07 0.00 0.10 0.15 0.20 0.30 8111 1
##
## Samples were drawn using NUTS(diag_e) at Sun Jan 25 18:41:15 2026.
## For each parameter, n_eff is a crude measure of effective sample size,
## and Rhat is the potential scale reduction factor on split chains (at
## convergence, Rhat=1).