fit_up = sampling(tsuga_unpooled, standat, iter=5000, refresh=0, chains=1)
fit_p = sampling(tsuga_pooled, standat, iter=5000, refresh=0, chains=1)
fit_pp = sampling(tsuga_ppool, standat, iter=5000, refresh=0, chains=1)
fit_ipp = sampling(tsuga_int_ppool, standat, iter=5000, refresh=0, chains=1)
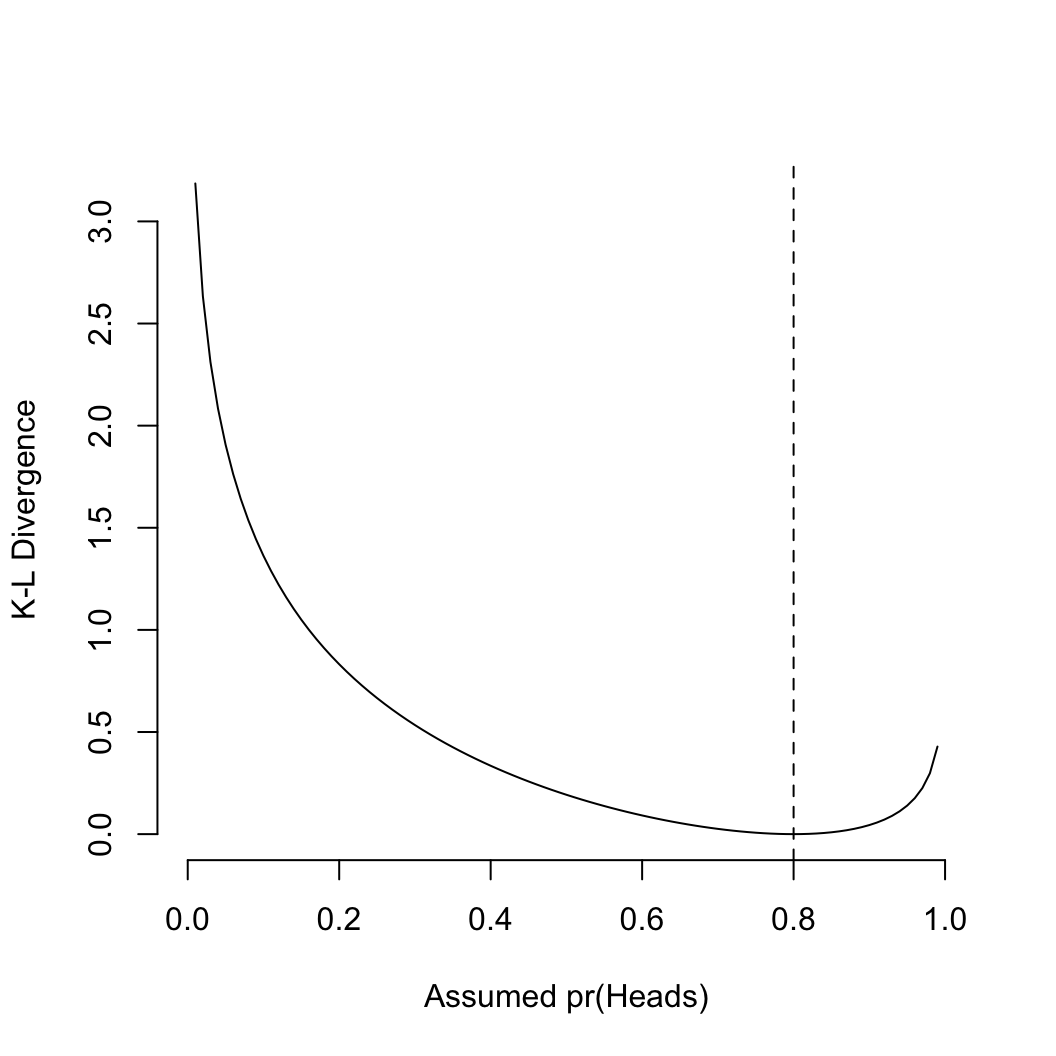
## note for the hierarchical model, a MAP is hard to find,
## here we use the min deviance as a quick and dirty estimate
## a better estimate is to compute the mean of all parameters, then
## compute the deviance with those values
pd = c(Pooled = mean(as.matrix(fit_p, pars="deviance")) - map_up$par["deviance"],
Unpooled = mean(as.matrix(fit_up, pars="deviance")) - map_p$par["deviance"],
Hierarchical = mean(as.matrix(fit_pp, pars="deviance")) - min(as.matrix(fit_pp, pars="deviance")),
Hierarchical_intercept = mean(as.matrix(fit_ipp, pars="deviance")) - min(as.matrix(fit_ipp, pars="deviance")))
dic = 1*pd + c(map_up$par["deviance"], map_p$par["deviance"], min(as.matrix(fit_pp, pars="deviance")),
min(as.matrix(fit_ipp, pars="deviance")))
dic = rbind(dic, dic - min(dic)); rownames(dic) = c("DIC", "delta")
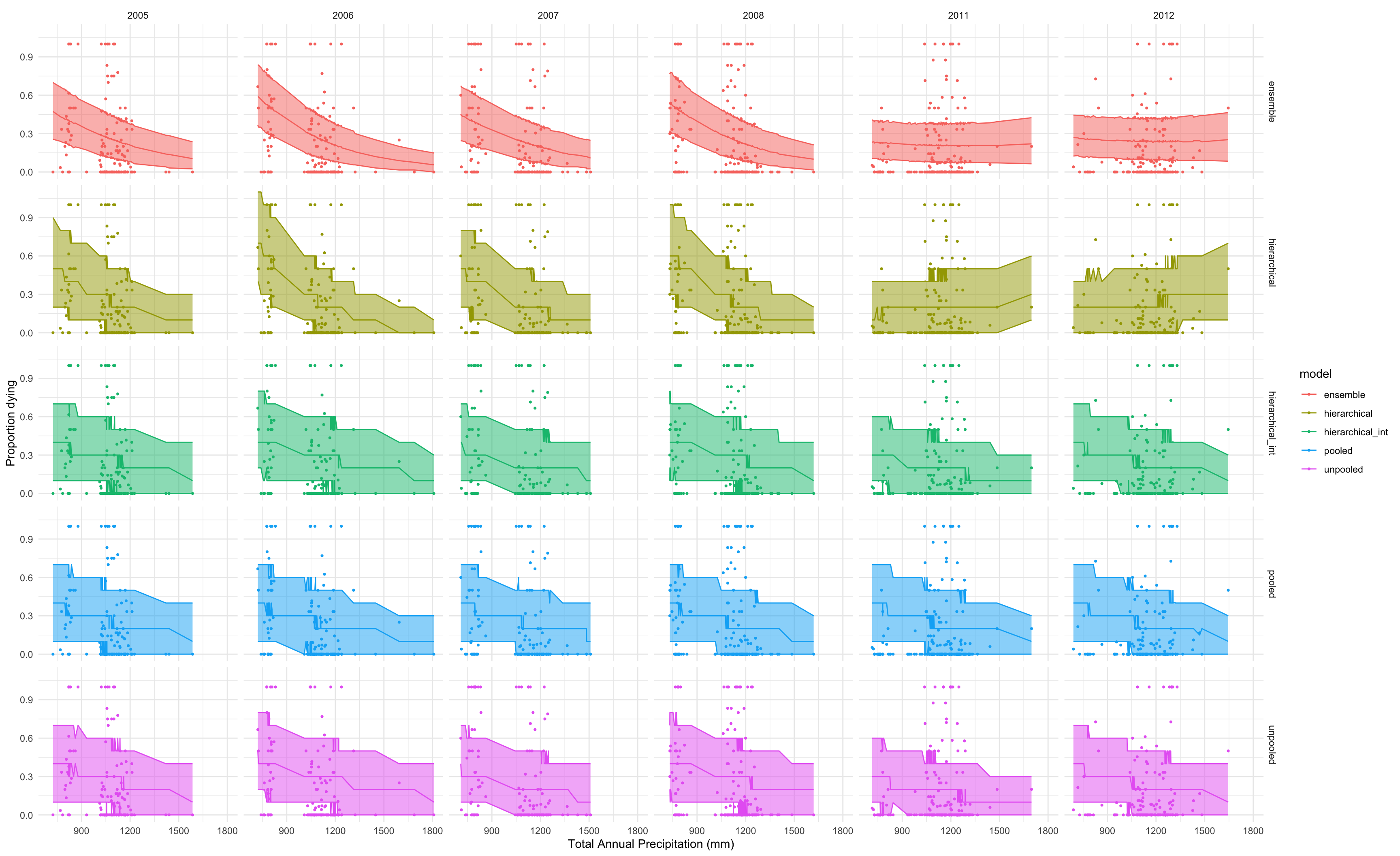
dic
## Pooled.deviance Unpooled.deviance Hierarchical Hierarchical_intercept
## DIC 5901.1156 5860.8207 5754.648 5872.4171
## delta 146.4677 106.1728 0.000 117.7692